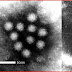
Coronaviruses are species in the genera of virus belonging to the subfamily Coronavirinae in the family Coronaviridae.
Coronaviruses are enveloped viruses with a positive-sense single-stranded RNA genome and a helical symmetry. The genomic size of coronaviruses ranges from approximately 16 to 31 kilobases, extraordinarily large for an RNA virus. The name "coronavirus" is derived from the Greek κορώνα, meaning crown, as the virus envelope appears under electron microscopy (E.M.) to be crowned by a characteristic ring of small bulbous structures. This morphology is actually formed by the viral spike (S) peplomers, which are proteins that populate the surface of the virus and determine host tropism. Coronaviruses are grouped in the order Nidovirales, named for the Latin nidus, meaning nest, as all viruses in this order produce a 3' co-terminal nested set of subgenomic mRNA's during infection.
Proteins that contribute to the overall structure of all coronaviruses are the spike (S), envelope (E), membrane (M) and nucleocapsid (N). In the specific case of SARS (see below), a defined receptor-binding domain on S mediates the attachment of the virus to its cellular receptor, angiotensin-converting enzyme 2 (ACE2). Members of the group 2 coronaviruses also have a shorter spike-like protein called hemagglutinin esterase (HE) encoded in their genome, but for some reason this protein is not always brought to expression (produced) in the cell.
Diseases of coronavirus
Coronaviruses primarily infect the upper respiratory and gastrointestinal tract of mammals and birds. Four to five different currently known strains of coronaviruses infect humans. The most publicized human coronavirus, SARS-CoV which causes SARS, has a unique pathogenesis because it causes both upper and lower respiratory tract infections and can also cause gastroenteritis. Coronaviruses are believed to cause a significant percentage of all common colds in human adults. Coronaviruses cause colds in humans primarily in the winter and early spring seasons. The significance and economic impact of coronaviruses as causative agents of the common cold are hard to assess because, unlike rhinoviruses (another common cold virus), human coronaviruses are difficult to grow in the laboratory.
Coronaviruses also cause a range of diseases in farm animals and domesticated pets, some of which can be serious and are a threat to the farming industry. Economically significant coronaviruses of farm animals include porcine coronavirus (transmissible gastroenteritis coronavirus, TGE) and bovine coronavirus, which both result in diarrhea in young animals. Feline Coronavirus: 2 forms, Feline enteric coronavirus is a pathogen of minor clinical significance, but spontaneous mutation of this virus can result in feline infectious peritonitis (FIP), a disease associated with high mortality. There are two types of canine coronavirus (CCoV), one that causes mild gastrointestinal disease and one that has been found to cause respiratory disease. Mouse hepatitis virus (MHV) is a coronavirus that causes an epidemic murine illness with high mortality, especially among colonies of laboratory mice. Prior to the discovery of SARS-CoV, MHV had been the best-studied coronavirus both in vivo and in vitro as well as at the molecular level. Some strains of MHV cause a progressive demyelinating encephalitis in mice which has been used as a murine model for multiple sclerosis. Significant research efforts have been focused on elucidating the viral pathogenesis of these animal coronaviruses, especially by virologists interested in veterinary and zoonotic diseases.
Replication
The infection cycle of coronavirus
Replication of Coronavirus begins with entry to the cell takes place in the cytoplasm in a membrane-protected microenvironment, upon entry to the cell the virus particle is uncoated and the RNA genome is deposited into the cytoplasm. The Coronavirus genome has a 5’ methylated cap and a 3’polyadenylated-A tail to make it look as much like the host RNA as possible. This also allows the RNA to attach to ribosomes for translation. Coronaviruses also have a protein known as a replicase encoded in its genome which allows the RNA viral genome to be transcribed into new RNA copies using the host cells machinery. The replicase is the first protein to be made as once the gene encoding the replicase is translated the translation is stopped by a stop codon. This is known as a nested transcript, where the transcript only encodes one gene- it is monocistronic. The RNA genome is replicated and a long polyprotein is formed, where all of the proteins are attached. Coronaviruses have a non-structural protein called a protease which is able to separate the proteins in the chain. This is a form of genetic economy for the virus allowing it to encode the most amounts of genes in a small amount of nucleotides.
Coronavirus transcription involves a discontinuous RNA synthesis (template switch) during the extension of a negative copy of the subgenomic mRNAs. Basepairing during transcription is a requirement. Coronavirus N protein is required for coronavirus RNA synthesis, and has RNA chaperone activity that may be involved in template switch. Both viral and cellular proteins are required for replication and transcription. Coronaviruses initiate translation by cap-dependent and cap-independent mechanisms. Cell macromolecular synthesis may be controlled after Coronavirus infection by locating some virus proteins in the host cell nucleus. Infection by different coronaviruses cause in the host alteration in the transcription and translation patterns, in the cell cycle, the cytoskeleton, apoptosis and coagulation pathways, inflammation, and immune and stress responses.
Severe acute respiratory syndrome
Main article: Severe acute respiratory syndrome
In 2003, following the outbreak of Severe acute respiratory syndrome (SARS) which had begun the prior year in Asia, and secondary cases elsewhere in the world, the World Health Organization issued a press release stating that a novel coronavirus identified by a number of laboratories was the causative agent for SARS. The virus was officially named the SARS coronavirus (SARS-CoV).
The SARS epidemic resulted in over 8000 infections, about 10% of which resulted in death. X-ray crystallography studies performed at the Advanced Light Source of Lawrence Berkeley National Laboratory have begun to give hope of a vaccine against the disease "since [the spike protein] appears to be recognized by the immune system of the host."
Recent discoveries of novel human coronaviruses
Following the high-profile publicity of SARS outbreaks, there has been a renewed interest in coronaviruses in the field of virology. For many years, scientists knew only about the existence of two human coronaviruses (HCoV-229E and HCoV-OC43). The discovery of SARS-CoV added another human coronavirus to the list. By the end of 2004, three independent research labs reported the discovery of a fourth human coronavirus. It has been named NL63, NL or the New Haven coronavirus by the different research groups. The naming of this fourth coronavirus is still a controversial issue, because the three labs are still battling over who actually discovered the virus first and hence earns the right to name the virus. Early in 2005, a research team at the University of Hong Kong reported finding a fifth human coronavirus in two pneumonia patients, and subsequently named it HKU1.
Species
Genus: Alphacoronavirus; type species: Alphacoronavirus 1
Species: Alphacoronavirus 1, Human coronavirus 229E, Human coronavirus NL63, Miniopterus Bat coronavirus 1, Miniopterus Bat coronavirus HKU8, Porcine epidemic diarrhea virus, Rhinolophus Bat coronavirus HKU2, Scotophilus Bat coronavirus 512
Genus Betacoronavirus; type species: Murine coronavirus
Species: Betacoronavirus 1, Human coronavirus HKU1, Murine coronavirus, Pipistrellus Bat coronavirus HKU5, Rousettus Bat coronavirus HKU9, Severe acute respiratory syndrome-related coronavirus, Tylonycteris Bat coronavirus HKU4
Genus Gammacoronavirus; type species: Avian coronavirus
Species: Avian coronavirus, Beluga whale coronavirus SW1
In April 2008, the following proposals were ratified by the ICTV:
2005.260V.04 To create the following species in the genus Coronavirus in the family Coronaviridae, named Goose coronavirus, Pigeon coronavirus, Duck coronavirus.
2006.009V.04 To create a species in the genus Coronavirus in the family Coronaviridae, named Human coronavirus NL63.
2006.010V.04 To create a species in the genus Coronavirus in the family Coronaviridae, named Human coronavirus HKU1.
2006.011V.04 To create a species in the genus Coronavirus in the family Coronaviridae, named Equine coronavirus.
In July 2009, the following proposals were ratified by the ICTV:
2008.085-122V.A.v3.Coronaviridae
2008.085V Create a new subfamily in the family Coronaviridae, order Nidovirales
2008.086V Name the new subfamily Coronavirinae
2008.087V Create a new genus in the proposed subfamily Coronavirinae
2008.088V Name the new genus Alphacoronavirus
2008.089V Assign three existing species (Human coronavirus 229E, Human coronavirus NL63, Porcine epidemic diarrhea virus) and five new species proposed in 2008.091-095V.01 to the proposed new genus Alphacoronavirus
2008.090V Designate proposed species Alphacoronavirus 1 as type species of the genus Alphacoronavirus
2008.091V Create new species named Alphacoronavirus 1 in the new genus
2008.092V Create new species named Rhinolophus bat coronavirus HKU2 in the new genus
2008.093V Create new species named Scotophilus bat coronavirus 512 in the new genus
2008.094V Create new species named Miniopterus bat coronavirus 1 in the new genus
2008.095V Create new species named Miniopterus bat coronavirus HKU8 in the new genus
2008.096V Create a new genus in the proposed subfamily Coronavirinae
2008.097V Name the new genus Betacoronavirus
2008.098V Assign the existing species Human coronavirus HKU1 and six new species proposed in
2008.100-105V.01 to the proposed genus Betacoronavirus
2008.099V Designate proposed species Murine coronavirus as type species of the genus Betacoronavirus
2008.108V Assign the two species proposed in 2008.110,111V.01 to the new genus
2008.109V Designate proposed species Avian coronavirus as type species of the new genus
2008.110V Create species named Avian coronavirus in the new genus
2008.111V Create species named Beluga whale coronavirus SW1 in the new genus
2008.112V Create a new subfamily in the family Coronaviridae, order Nidovirales
2008.113V Name the new subfamily Torovirinae
2008.114V Create a new genus in the subfamily Torovirinae
2008.115V Name the new genus Bafinivirus
2008.116V Assign the species White breamVirus (proposed in 2008.118V.01) to the new genus
2008.117V Designate species White bream virus as type species in the new genus
2008.118V Create species named White bream virus in the new genus
2008.119V Remove the genus Torovirus from the family Coronaviridae
2008.120V Reassign the genus Torovirus to the subfamily Torovirinae
2008.121V.U Remove (abolish) 18 species (Human enteric coronavirus, Human coronavirus OC43, Bovine coronavirus, Porcine hemagglutinating encephalomyelitis virus, Equine coronavirus, Murine hepatitis virus, Puffinosis coronavirus, Rat coronavirus, Transmissible gastroenteritis virus, Canine coronavirus, Feline coronavirus, Infectious bronchitis virus, Duck coronavirus, Goose coronavirus, Pheasant coronavirus, Pigeon coronavirus, Turkey coronavirus, Severe acute respiratory syndrome coronavirus) from the genus Coronavirus
2008.122V.U Reassign species Human coronavirus 229E, Human coronavirus NL63 and Porcine epidemic diarrhea virus to the new genus Alphacoronavirus and Human coronavirus HKU1 to the new genus Betacoronavirus