Sputnik virophage (from Russian cпутник "satellite", Latin "virus" and Greek φάγειν phagein "to eat") is a subviral agent that reproduces in amoeba cells that are already infected by a certain helper virus; Sputnik uses the helper virus's machinery for reproduction and inhibits replication of the helper virus.
Viruses like Sputnik that depend on co-infection of the host cell by helper viruses are known as satellite viruses. At its discovery in a Paris water-cooling tower in 2008 Sputnik was the first known satellite virus that inhibited replication of its helper virus and thus acted as a parasite of that virus. In analogy to the term bacteriophage it was called a virophage.
Virology
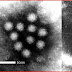
Sputnik virophage is icosahedral in shape and 50 nanometres in size. It has been found to multiply inside of Acanthamoeba species, but only if that amoeba is infected with the large mamavirus. Sputnik harnesses the mamavirus proteins to rapidly produce new copies of itself.
Mamavirus is formally known as Acanthamoeba polyphaga mimivirus (APMV) and is a close relative of the previously known mimivirus. The mimivirus is a giant in the viral world; it has more genes than many bacteria and performs functions that normally occur only in cellular organisms. The mamavirus is even larger than the mimivirus, but the two are very similar in that they form large viral factories and complex viral particles.[2] Virophage growth is deleterious to APMV and results in the production of abortive forms and abnormal capsid assembly of APMV. In one of the experiments done by inoculating Acanthamoeba polyphaga with water containing an original strain of APMV, it was discovered that several capsid layers accumulate unsymmetrically on one side of the viral particle causing the virus to become ineffective. Sputnik decreased the yield of infective viral particle by 70% and also reduced the amoeba lysis by threefold at 24h.
Sputnik has a circular double stranded DNA genome consisting of 18,343 base pairs.[2] It contains genes able to infect all three domains of life: Eukarya, Archaea and Bacteria. Of the twenty-one predicted protein-coding genes, three are apparently derived from APMV itself, one is a homologue of an archaeal virus, and four others are homologues of proteins in bacteriophages and eukaryotic viruses. Thirteen are ORFans, that is they do not have any detectable homologues in current sequence databases. The Sputnik genome has a high A + T content (73%) similar to that of APMV.
Several other homologues such as those of a primase–helicase, a packaging ATPase, an insertion sequence transposase DNA-binding subunit, and a Zn-ribbon protein, were detected in the Global Ocean Survey environmental data set, suggesting that virophages could be a currently unknown family of viruses.
Sputnik was found to contain genes that were shared by APMV. These genes could have been acquired by Sputnik after the association of APMV with the host and then interaction between the virophage and the viral host. Recombination within the viral factory might have resulted in the exchange of genes. Sputnik is one of the most convincing pieces of evidence for gene mixing and matching between viruses.
The presence of these genes homologous to the mimivirus in Sputnik suggests that gene transfer between Sputnik and the mimivirus can occur during the infection of Acanthamoeba. Therefore, it is hypothesized that the virophage could be a source of vehicle mediating lateral gene transfer between giant viruses, which constitute a significant part of the DNA virus population in the marine environments. Moreover, the presence of three APMV genes in Sputnik implies that gene transfer between a virophage and a giant virus is crucial to viral evolution.
In March 2011, two additional virophages were described: the Mavirus virophage which preys on the giant Cafeteria roenbergensis virus, and the Organic Lake Virophage, found in the salty Organic Lake in Antarctica, and which preys on viruses that attack algae. All host viruses of the known virophages belong to the group of nucleocytoplasmic large DNA viruses.
Viruses like Sputnik that depend on co-infection of the host cell by helper viruses are known as satellite viruses. At its discovery in a Paris water-cooling tower in 2008 Sputnik was the first known satellite virus that inhibited replication of its helper virus and thus acted as a parasite of that virus. In analogy to the term bacteriophage it was called a virophage.
Virology
Sputnik virophage is icosahedral in shape and 50 nanometres in size. It has been found to multiply inside of Acanthamoeba species, but only if that amoeba is infected with the large mamavirus. Sputnik harnesses the mamavirus proteins to rapidly produce new copies of itself.
Mamavirus is formally known as Acanthamoeba polyphaga mimivirus (APMV) and is a close relative of the previously known mimivirus. The mimivirus is a giant in the viral world; it has more genes than many bacteria and performs functions that normally occur only in cellular organisms. The mamavirus is even larger than the mimivirus, but the two are very similar in that they form large viral factories and complex viral particles.[2] Virophage growth is deleterious to APMV and results in the production of abortive forms and abnormal capsid assembly of APMV. In one of the experiments done by inoculating Acanthamoeba polyphaga with water containing an original strain of APMV, it was discovered that several capsid layers accumulate unsymmetrically on one side of the viral particle causing the virus to become ineffective. Sputnik decreased the yield of infective viral particle by 70% and also reduced the amoeba lysis by threefold at 24h.
Sputnik has a circular double stranded DNA genome consisting of 18,343 base pairs.[2] It contains genes able to infect all three domains of life: Eukarya, Archaea and Bacteria. Of the twenty-one predicted protein-coding genes, three are apparently derived from APMV itself, one is a homologue of an archaeal virus, and four others are homologues of proteins in bacteriophages and eukaryotic viruses. Thirteen are ORFans, that is they do not have any detectable homologues in current sequence databases. The Sputnik genome has a high A + T content (73%) similar to that of APMV.
Several other homologues such as those of a primase–helicase, a packaging ATPase, an insertion sequence transposase DNA-binding subunit, and a Zn-ribbon protein, were detected in the Global Ocean Survey environmental data set, suggesting that virophages could be a currently unknown family of viruses.
Sputnik was found to contain genes that were shared by APMV. These genes could have been acquired by Sputnik after the association of APMV with the host and then interaction between the virophage and the viral host. Recombination within the viral factory might have resulted in the exchange of genes. Sputnik is one of the most convincing pieces of evidence for gene mixing and matching between viruses.
The presence of these genes homologous to the mimivirus in Sputnik suggests that gene transfer between Sputnik and the mimivirus can occur during the infection of Acanthamoeba. Therefore, it is hypothesized that the virophage could be a source of vehicle mediating lateral gene transfer between giant viruses, which constitute a significant part of the DNA virus population in the marine environments. Moreover, the presence of three APMV genes in Sputnik implies that gene transfer between a virophage and a giant virus is crucial to viral evolution.
In March 2011, two additional virophages were described: the Mavirus virophage which preys on the giant Cafeteria roenbergensis virus, and the Organic Lake Virophage, found in the salty Organic Lake in Antarctica, and which preys on viruses that attack algae. All host viruses of the known virophages belong to the group of nucleocytoplasmic large DNA viruses.













0 komentar:
Posting Komentar